Help
ECOdrug: a database connecting drugs and conservation of their targets across species
Bas Verbruggen, Lina Gunnarsson, Erik Kristiansson, Tobias Österlund, Stewart F. Owen, Jason R. Snape, and Charles R. Tyler
Nucleic Acids Research, gkx1024
1. Downloads
2. Methods
3. Drug page
4. Drug target page
5. Contact us
Downloads
Majority vote:
Targets
Drugs & Targets
Ensembl:
Targets
Drugs & Targets
EggNOG:
Targets
Drugs & Targets
InParanoid:
Targets
Drugs & Targets
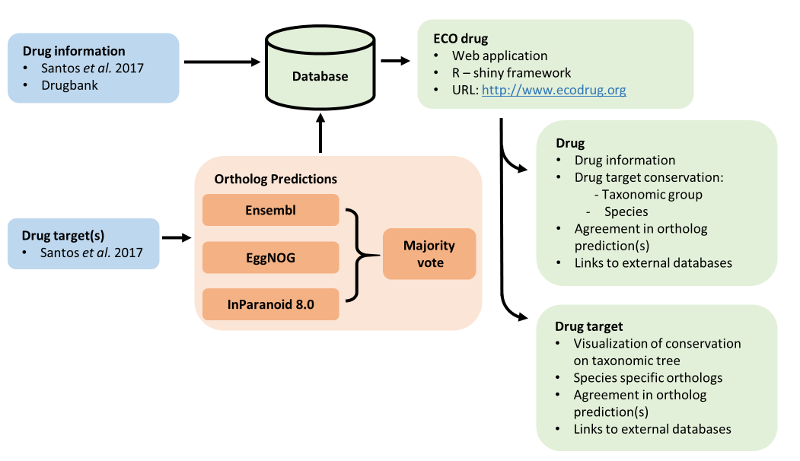
Methods
Drug targets
A comprehensive dataset of APIs and corresponding drug targets was assembled by Santos et al. 2017 [1].
ECOdrug focuses on conservation of human drug targets, therefore any non-human drug targets
(e.g. bacterial or viral proteins) were removed from the dataset. Proteins in this dataset were defined
by their Uniprot identifier. We used biomaRt to retrieve any associated ensemble gene and ensemble protein
identifiers.
Ortholog prediction
Drug target ortholog predictions were derived from three prediction methods. To improve accuracy we
combined results of these methods together where possible. The three chosen databases were:
Ensembl, EggNOG and InParanoid.
Ensembl:
Orthologs of drug targets from Santos et al. were retrieved through biomaRt by mapping ensembl gene IDs
to all available ensembl homology attributes (i.e. the species in the Ensembl Compara database)(Febuary 2017).
Beyond chordate spaces the ortholog predictions had to be retrieved from Ensembl panCompara [6] as Ensembl Compara
only contains two invertebrates and no plants or green algae. Ortholog predictions for these species were retrieved
from panEnsembl release 30.
EggNOG:
Ortholog predictions of human drug targets, defined by ensembl peptide IDs, are retrieved by iterating
over EggNOGs taxonomic levels from closest to Homo sapiens (Hominidae NOG) to furthest (Eukaryotes NOG).
At every level drug target ortholog are retrieved for species represented in the group. These species
are ignored at subsequent taxonomic levels so orthologs are only retrieved from the closest possible
taxonomic level.
InParanoid:
The standalone version of InParanoid was used to obtain up-to-date ortholog predictions for drug targets
by running the UniProt Homo sapiens reference proteome (UP000005640_9606.fasta; UniProt Release 2016_04,
Ensembl release 84 and Ensembl Genome release 31) against the reference proteomes of other species in
the Uniprot database.
Combining ortholog predictions:
Ortholog predictions from Ensembl, EggNOG and InParanoid each represented a different set of species.
However there are species that are shared between databases. In such cases we compared and combined
the presence/absence of orthologs to increase the confidence in ortholog prediction. For species that
were available in all three methods we applied a majority vote principle, i.e. two or more databases
have to agree on the presence/absence of a drug target ortholog in a species. This principle does
not carry to situations where a species is represented in only two methods, in such cases the majority vote
indicates presence of an ortholog if at least one database predicts an ortholog.
The combined presence/absence predictions and individual method results are available through this application
written in the R-shiny framework.
References:
[1] Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, Karlsson A, Al-Lazikani B,
Hersey A, Oprea TI
et al
.:
A comprehensive map of molecular drug targets.
Nat Rev Drug Discov 2017, 16(1):19-34.
Drug page
The drug page is a page on which general information on drugs,
an overview of their targets and conservation of those targets can be retrieved.
This information will be displayed once a drug is selected from the drug selector. Drugs can
be found by typing in their name in the field or by selecting it from the alfabetic list.
Having selected a drug, the main panel displays information on the selected drug.
At the top you will find generic information on the drug, as derived from DrugBank [1].
Depending on availability, the drug page should displays the following information about the drug:
- Drug name
- Drug synonyms, alternative names used for this drug, are displayed below the name
- Description of the drug
- Drugbank identifier (+ direct link to drugbank website)
- Type of molecule (e.g. small molecule or biological)
- Target species (usually Humans)
- Status of the drug according to drugbank
- ATC code, indicating therapeutic area for the drug [2]
- Mode of Action: A description of the mechanism of action of the drug, derived from Drugbank
Following the description of the drug, an overview of target conservation is shown.
Drug targets are derived from Santos
et al
. 2017 [3], and ortholog predictions identified as
described in the method section on this page.
Depending on the chosen display table one of two tables is shown.
Taxonomic group
:
This table provides a high level overview of ortholog predictions for all drug targets
associated with the selected drug. The rows in the table indicate taxonomic groups and
a seperate column is shown for every drug target. Values in the table indicate the number
of species for which an ortholog to the drug target was identified according to a majority
vote of Ensembl, EggNOG and InParanoid compared to the total number of species belonging to
that particular taxonomic group. A colour code from red to green indicates how strong predicted
conservation is. The species included in this table can either be all the species shared
amongst the three databases, or include all species. This depends on whether the checkbox
for 'only shared species for phyla' is checked or not.
Species
:
This table provides species specific orhtolog predictions for the drug targets associated with
the selected drug. In the settings panel a choice can be made to limit the number of species to
a certain taxonomic group. Selecting 'Eukaryota' will display all available species. By default
ortholog predictions for the majority vote approach are shown.
TRUE
indicates presence of an ortholog,
FALSE
indicates absence. It is possible to display predictions from an individual prediction method by
selecting it in the settings panel. In this case predicted orthologs are shown by their identifiers and
links to external databases.
Every drug target has a button with its UniProt ID that leads to its corresponding Drug Target page.
A download button is present to download the displayed table to a .csv file.
References:
[1] Drugbank:
https://www.drugbank.ca/
[2] ATC codes:
https://www.whocc.no/atc_ddd_index/
[3] Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, Karlsson A, Al-Lazikani B,
Hersey A, Oprea TI
et al
.:
A comprehensive map of molecular drug targets.
Nat Rev Drug Discov 2017, 16(1):19-34.
Drug target page
The drug target page is a page through which conservation of drug targets can be retrieved
Available information will be displayed once a target is selected from the target selector. Targets can
be found by typing in their name in the field or by selecting it from the alfabetic list.
Available identifiers are: HGNC, Ensembl or UniProt identifiers.
Having selected a target, the main panel displays brief description of the target and links to external
databases. Below the links there is a list of drugs that are associated with the selected drug target.
The buttons direct the user to the corresponding drug page.
Following the general target information is a table that contains ortholog predictions.
Results are displayed for the chosen taxonomy range (see settings panel), choosing 'Eukaryota' provides all
available species.
The table contains information on the species (latin name, common name and NCBI taxonomy identifier), the
number of databases containing the species, the majority vote ortholog prediction and predictions from
individual prediction methods (EggNOG, Ensembl and InParanoid).
Presence/absence of an ortholog is indicated by
TRUE
/
FALSE
for the majority vote results,
for the individual databases identifiers (+ links) are provided.
The table allows sorting based on the column names and has an associated search function.
A download button is present to download the displayed table to a .csv file.
Contact us
Dr. Bas Verbruggen: [email protected]
Dr. Lina Gunnarsson: [email protected]
Prof. Charles Tyler: [email protected]
